Section: New Results
Mathematical and Mechanical modeling
A 3D contact-mechanics model of the heart and thorax for seismocardiography
Participants : Alexandre Laurin [correspondant] , Sébastien Imperiale, Dominique Chapelle, Philippe Moireau.
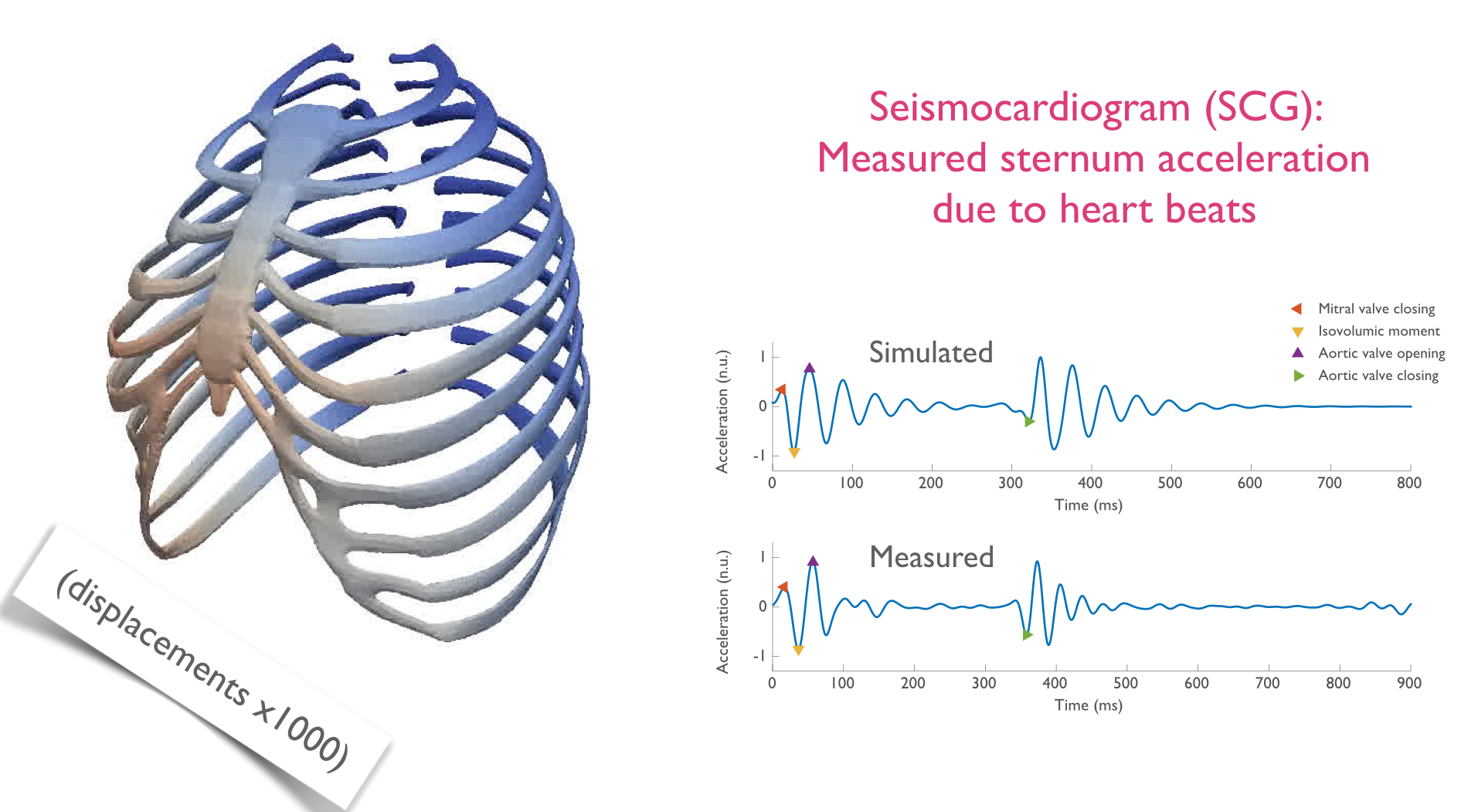
The current interpretation of seismocardiogram fiducial points depends on their phenomenological association with the timing of events on simultaneous echocardiograms. Signal processing methods can be devised to acquire these timings automatically (see [21] and [22]). So far, no causal framework has been tested to explain this timing, nor their direction and amplitude. This work attempted to adapt a comprehensive 3D cardiac model to interact through contact with a model of the thoracic cage. The heart model was designed to represent multi-scale, multi-physics physiological processes such as the electrical activation, the mechanical contraction, as well as the system circulation. The objective is to link observed acceleration of the sternum to the underlying physiology, and offer a potential mechanical explanation for them. The modelling chain necessary to go from the heart model to a simulated SCG has been successfully implemented (see Figure 1). Furthermore, the complexity of the thoracic model has been substantially reduced, without deteriorating results, to improve the portability of the entire process. Once the relevant parameters of in-vivo thoraces will have been precisely identified, it will be possible to compute heart forces and the various cardiac events that cause them directly from SCG measurements. The subsequent aim is to apply the model to ageing and pathological physiologies.
|
Multi-scale modeling of muscle contraction
Participants : François Kimmig [correspondant] , Dominique Chapelle, Matthieu Caruel.
This works aims at proposing a multi-scale model of the muscular contraction that can be used in the context of cardiac simulation. The modeling will be based on the stochastic equations that describe muscular contraction at the molecular level. Asymptotic counterparts of the stochastic model will be considered in order to provide pertinent simplified models. The modeling elements will be confronted with experiments that will be performed on cardiac muscle cells by collaborators in the team of Professor Vincenzo Lombardi at the University of Florence.
In the framework of this collaboration, a chemicomechanical model is being implemented into CardiacLab, a simulation environment developed by the team. It will enrich the range of modeling tools of the team for the active contribution of muscle cells to the cardiac behavior.
Multiphysics and multiscale modelling, data-model fusion and integration of organ physiology in the clinic: ventricular cardiac mechanics
Participants : Radomir Chabiniok [correspondant] , Philippe Moireau, Dominique Chapelle, Maxime Sermesant [Team Asclepios] .
With heart and cardiovascular diseases continually challenging healthcare systems worldwide, translating basic research on cardiac (patho)physiology into clinical care is essential. Exacerbating this already extensive challenge is the complexity of the heart, relying on its hierarchical structure and function to maintain cardiovascular flow. Computational modelling has been proposed and actively pursued as a tool for accelerating research and translation. Allowing exploration of the relationships between physics, multiscale mechanisms and function, computational modelling provides a platform for improving our understanding of the heart. Further integration of experimental and clinical data through data assimilation and parameter estimation techniques is bringing computational models closer to use in routine clinical practice. This work published in [17] reviews developments in computational cardiac modelling and how their integration with medical imaging data is providing new pathways for translational cardiac modelling.
Eidolon: visualization and computational framework for multi-modal biomedical data analysis
Participant : Radomir Chabiniok [correspondant] .
Biomedical research, combining multi-modal image and geometry data, presents unique challenges for data visualization, processing, and quantitative analysis. Medical imaging provides rich information, from anatomical to deformation, but extracting this to a coherent picture across image modalities with preserved quality is not trivial. Addressing these challenges and integrating visualization with image and quantitative analysis results in Eidolon, a platform which can adapt to rapidly changing research workflows. In the paper [26] we outline Eidolon, a software environment aimed at addressing these challenges, and discuss the novel integration of visualization and analysis components. These capabilities are demonstrated through the example of cardiac strain analysis, showing the Eidolon supports and enhances the workflow.
Mathematical and numerical modeling of elastic waves propagation in the heart
Participants : Federica Caforio [correspondant] , Dominique Chapelle, Sébastien Imperiale.
The objective of this work is to develop a rigorous mathematical and numerical background for the extension and dissemination of elastography imaging modalities, applied to the cardiac setting. The problems treated concern the topics of mathematical modelling, numerical analysis and scientific computing. More precisely, the plan is to define a linearised model for the propagation of elastic waves in the heart, to study approximations of these models and define adapted numerical methods for the discretisation of the resulting partial differential equations.